Getting Started
What is HNN?
HNN simulates primary current dipoles, i.e. the neural sources of the sensor-level EEG/MEG signal. The core of HNN is a preconstructed large-scale biophysically detailed template model of a neocortical column model that can be synaptically activated with exogenous layer-specific thalamocortical and cortical-cortical input. HNNs template model is based on generalizable principles of neocortical circuitry and contains pyramidal and inhibitory across the cortical layers (see Figs 1 & 2, below). HNN simulates the primary electrical current source representing a current dipole in source localized EEG/MEG signals based on their biophysical origin from the intracellular current flow in the pyramidal neuron dendrites, along with the multiscale corresponding cell and circuit level activity. As such, HNN provides a bidirectional hypothesis testing and development tool aimed to bridge localized macroscale human EEG/MEG signals to the underlying micro/meso scale activity, which can be interrogated in animal models (Fig 1). Importantly, HNN is designed to study the cell and circuit-level origin of waveforms from a single localized region of the cortex. It does not provide tools for the source localization estimation process (i.e. inverse modeling). There are many excellent source localization software for this estimation process. For an example of how to perform source localization using MNE-python followed by cell and circuit level interpretation with HNN, see our example in HNN-Core.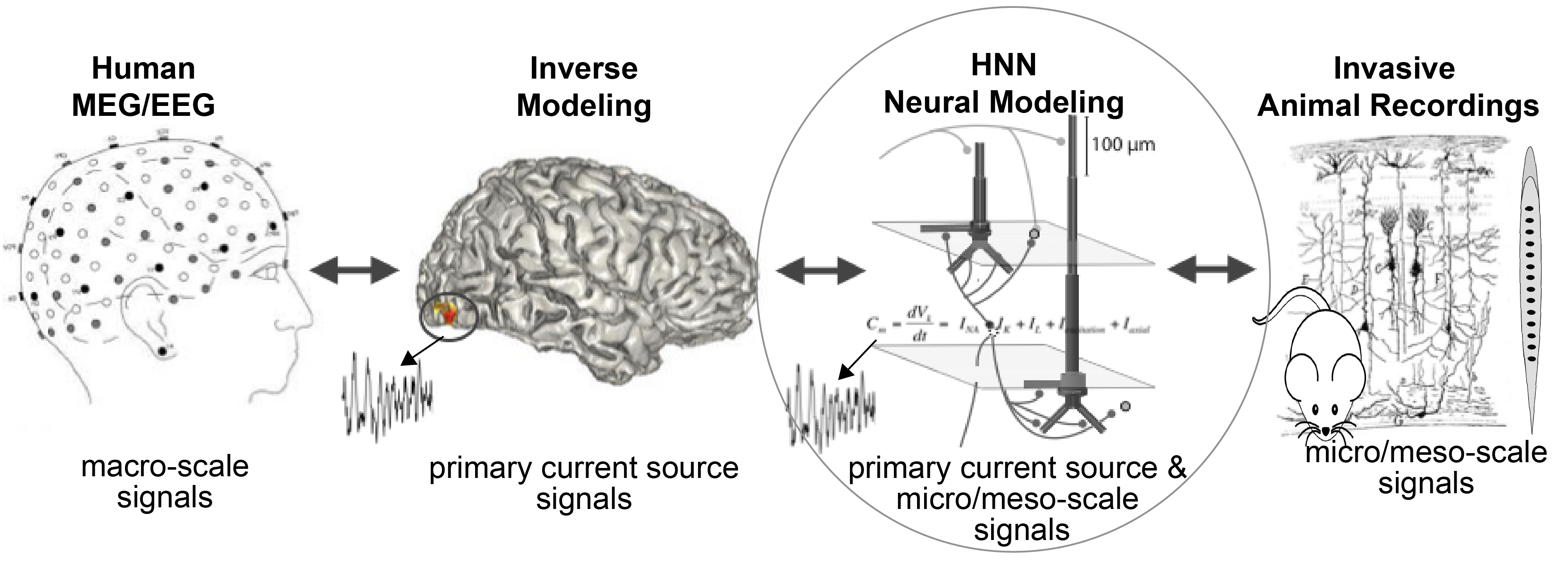
Figure 1
HNN’s template model has been used to study the origin of some of the most measured EEG/MEG signals including event-related potentials, low-frequency rhythms, and changes in these signals with behavior, neuropathology, and non-invasive brain stimulation (see publications). To facilitate the use of this complex neural model by a wide user base, HNN’s template model is embedded in a graphical user interface (HNN-GUI) and developed workflows and tutorials to teach users how to interact with the model and visualize the multi-scale signals underpinnings of source localized MEG/EEG signals. Due to the level of biophysical detail in HNN’s model, the current dipole signal generated in HNN can be directly and statistically compared to the source localized MEG/EEG signal. Out of the box, users can quickly and easily begin to develop and test hypotheses on the origin of their data. Through an interactive GUI investigation users can visualize the impact of changes in model parameters on current dipole signals along with the underlying changes in multilayer cell and circuit dynamics. Once a good fit to the recorded M/EEG data is found, the model provides detailed and targeted predictions on underlying cell and circuit activity that can be tested with invasive recordings or other imaging modalities (see for example Khan et al 2015, Sherman et al 2016, Bonaiuto et al 2021).
Our HNN-GUI tutorials are designed to teach users how to begin to study the origin of ERP and brain rhythms based on our prior published studies. We provide data and initial parameter sets along with workflows to teach users how to do what we did to develop and test hypotheses on the neural origin of these signals. Once this workflow is learned, users will be able to use HNN to develop and test their own hypotheses. Our goal is not to promote singular theories on the origin of any particular signal, but rather to provide a tool based on known biophysics and training resources where researchers can develop testable predictions on the neural origin of their data. You can read more on the process of modeling large-scale neural models on our HNN-GUI tutorial page.

HNN-Core: HNN’s Computational Core is Developed in NEURON-Python and Designed with Best Open-Source Software Design Practice for Modularity and Expansion
HNN-Core is a Python package designed to have all the functionality of the HNN-GUI in a command line interface. HNN-core is developed using best practices in open-source software design to facilitate expansion in its utility and community development. Tutorials of use that mimic the HNN-GUI tutorials are available, see the HNN-Core tutorials page and further functionality are available, see examples page on Github. In practice, after learning how to use HNN through the GUI, if you know a bit about computational neuroscience and Python coding, we encourage you to use HNN-Core and contribute to its development. Please see our contributing guidelines and issues page on our HNN-Core github page. Our hope is that, with a large enough HNN user and developer base, crowdsourced knowledge will lead to new testable discoveries on the biophysical origin of human EEG/MEG signals and treatments for neuropathology.
Moving Beyond the HNN Template Model
We are still in the early days of understanding the contributors to macroscale human EEG/MEG signals. HNN’s template model and workflows of use are designed as a starting point for developing and testing predictions on the detailed neural mechanisms contributing to EEG/MEG signals based on the known biophysics of their generation. The multi-scale simulation (layer and cell-specific spiking, LFP/CSD, etc..) possible with HNN facilitates testing of model-derived predictions across multiple recordings scales and species. HNN-core is designed in a modular fashion so that it can be easily expanded as new knowledge of the cells and circuits that contribute to these signals is learned. A goal is to be able to use other neocortical models within the HNN workflows. Currently, there are 3 cortical "template" models available to study in HNN-Core. We are in the process of developing a new GUI using HNN-Core functions with all of the original GUI functionality and more. Once the new HNN-core-based GUI is complete and documented, the original HNN-GUI will be deprecated. Additional functionality in HNN-core will include the ability to calculate and visualize LFP/CSD (currently possible in HNN-Core) and intracellular calcium signals, more advanced parameter estimation techniques, and examples of how to integrate source localization in MNE-Python with circuit-level interpretation in HNN.
HNN’s Neocortical Column Template Model & The Biophysical Origin of Source Localize MEG/EEG Primary Currents (i.e. Current Dipoles)
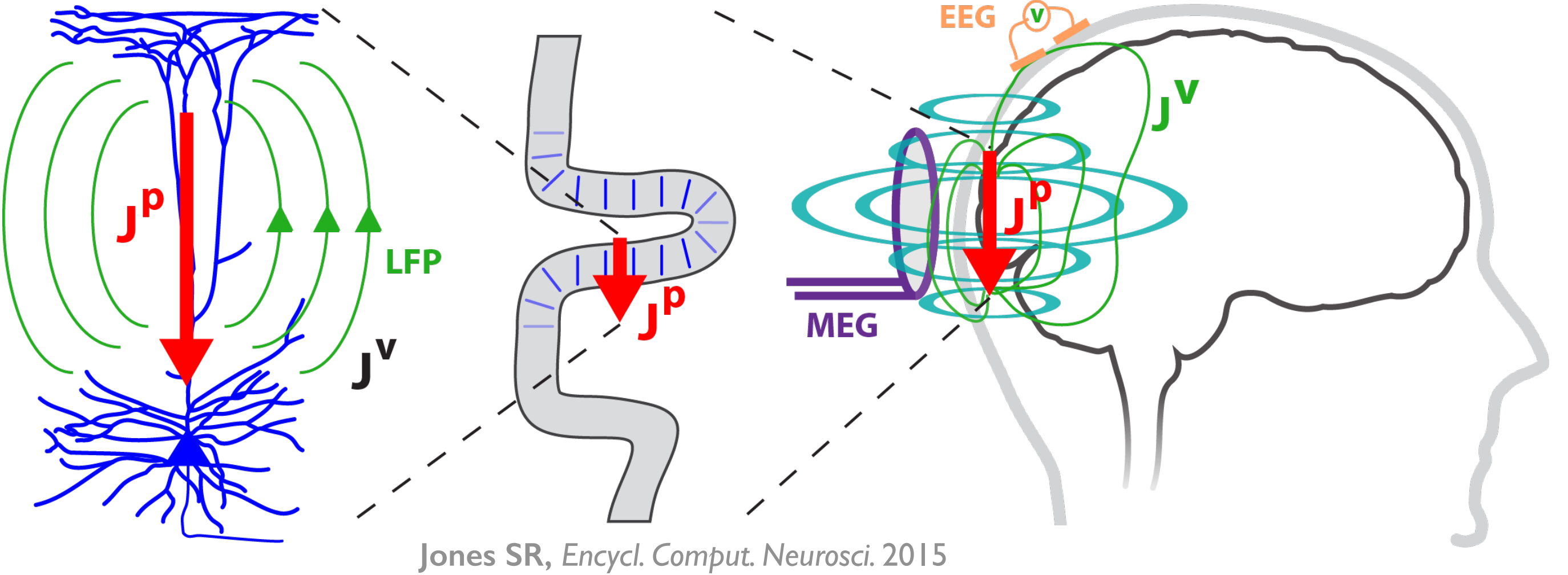
Figure 2
HNN’s underlying neural model simulates the primary electrical currents in the neocortex that create EEG/MEG signals. The model simulations are based on the biophysical origins of the primary electrical currents (see Figure 2). The primary electrical currents (JP) are assumed to be generated by the post-synaptic, intracellular longitudinal current flow in the long and spatially aligned dendrites of a large population of synchronous neocortical pyramidal neurons1,2. These primary currents create volume currents (JV) that propagate through the brain tissue, cerebral spinal fluid, and skull. It is the volume currents that are pick up by the EEG/MEG sensors. “Inverse solution methods” (i.e., source localization methods) applied to the sensor data estimate the location, timing, and directionality of the primary electrical current dipole sources (Jp) (i.e., equivalent current dipoles1).
| HNN simulates these primary currents from a canonical model of a layered neocortical column via the net intracellular electrical current flow in the pyramidal neuron dendrites (see schematic to the right. Further details can be found at our HNN Template Model page). The units of measure that come out of the model are directly comparable to those estimated from source localization methods, namely, nano-ampere-meters (nAm). |

|
Building from a history of prior published work, a pre-assembled and pre-tuned neocortical template model that represents a generalizable, canonical cortical column is provided. The template circuit includes a scalable number of neurons and adjustable parameters along with simulation tools to perturb the local network with layer- and cell-specific external drive (i.e., spike driven synaptic input and/or tonic applied currents). See details in HNN Template Model . Tools are also provided for users to directly compare simulated electrical sources with recorded source localized data*.
Importantly, HNN was created to be a hypothesis development and testing tool. While the underlying neural model is biophysically principled and based on generalizable features of cortical networks, and also includes neurons with realistic physiology and morphology (see details in HNN Template Model), the model is nonetheless a reduced model of neocortical network dynamics. Any conclusions made are based on the underlying model assumptions, which are outlined in detail in our HNN Template Model page. Users should become familiar with these assumptions at the start of using HNN. We have found the model to be useful for generating novel and testable hypotheses on the circuit origin of some of the most commonly measured signals, including ERPs and low-frequency oscillations3-7, which provide the basis for the tutorials. As discussed above, the modular design in HNN-core facilitates the process of using other more specialized neocortical column models in HNN. We are working toward a framework to use neocortical column models developed in other software, e.g. (http://www.neurosimlab.com/netpyne) into the HNN simulation framework. We are open-source and encourage input and expansion from the community (see Users Forum).
*If you only have sensor data, you can still benefit from our software. The primary currents are the foundation of the sensor signal and, as such, will have similar activity profiles.
