Location: 164 Angell St.
Floor 4
Innovation Zone
Date: May 23, 2019
Time: 2 PM – 4:30 PM
Speaker:
Salvador Dura-Bernal, Ph.D.
Research Assistant Professor
State University of New York, Downstate Medical Center
Sign Up:
https://forms.gle/nuwBxw7H1LdAhBrm9
Note that sign up is required for this event. The workshop is limited to 40 people, and we will check to ensure that the attendee information matches the information in the sign-up form above
Agenda
2:00 – 2:45 PM: Introduction to multi-scale modeling using NetPyNE and example applications (Seminar-like)
2:45 – 4:30 PM: Hands-on tutorial covering tool installation, building a simple and a complex network, running simulations, visualizing and analyzing network and simulations results, and using the GUI.
Pre-reqs:
Basic neurobiology (e.g., working knowledge of neurons, dendrites, ionic channels, voltage, synapses, etc.) is recommended
Basic programming skills are recommended but not strictly required. GUI examples in particular do not require any additional expertises.
Workshop Overview
We are in the process of converting HNN to be compatible with models built in NetPyNe (www.netpyne.org) so that individualized network models can be easily and flexibly developed and used in the HNN environment. Please join us in a workshop to learn more about NetPyNe. Click here to be directed to eLife’s most recent publication on NetPyNE.
——————————
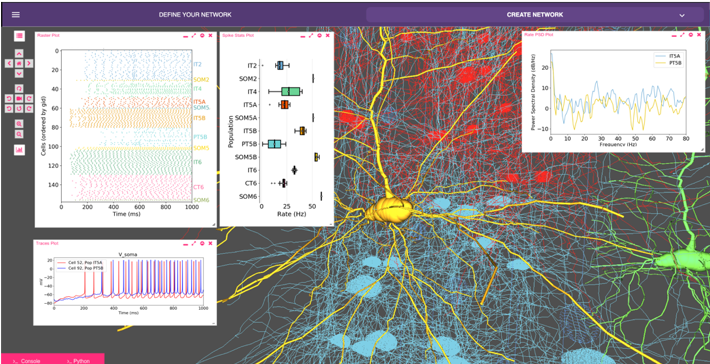
Biophysical modeling of neuronal networks helps to integrate and interpret rapidly growing and disparate experimental datasets at multiple scales. The NetPyNE tool (www.netpyne.org) provides both programmatic and graphical interfaces to develop data-driven multiscale network models in NEURON.
NetPyNE clearly separates model parameters from implementation code. Users provide specifications at a high level via a standardized declarative language, eg. connectivity rules, to create millions of cell-to-cell connections.
NetPyNE then enables users to generate the NEURON network, run efficiently parallelized simulations, optimize and explore network parameters through automated batch runs, and use built-in functions for visualization and analysis — connectivity matrices, voltage traces, spike raster plots, local field potentials, and information theoretic measures. NetPyNE also facilitates model sharing by exporting and importing standardized formats (NeuroML and SONATA).
NetPyNE is already being used in over 20 labs to teach computational neuroscience students and by modelers to investigate brain regions and phenomena, including cortical circuits, hippocampus, cerebellum, schizophrenia, epilepsy and neurostimulation. It is also being integrated with the Human Neocortical Neurosolver tool (hnn.brown.edu) developed at Brown.
——————————
This series is organized by the Providence VA Center for Neurorestoration and Neurotechnology Core for Recording, Decoding and Computational Neuroscience, Brown University Center for Central Nervous System Function, and the Carney Institute.

