
HUMAN NEOCORTICAL NEUROSOLVER
An open-source software for developing and testing hypotheses on the cellular- and circuit-level mechanisms of electro- and magneto-encephalography (EEG/MEG) signals.

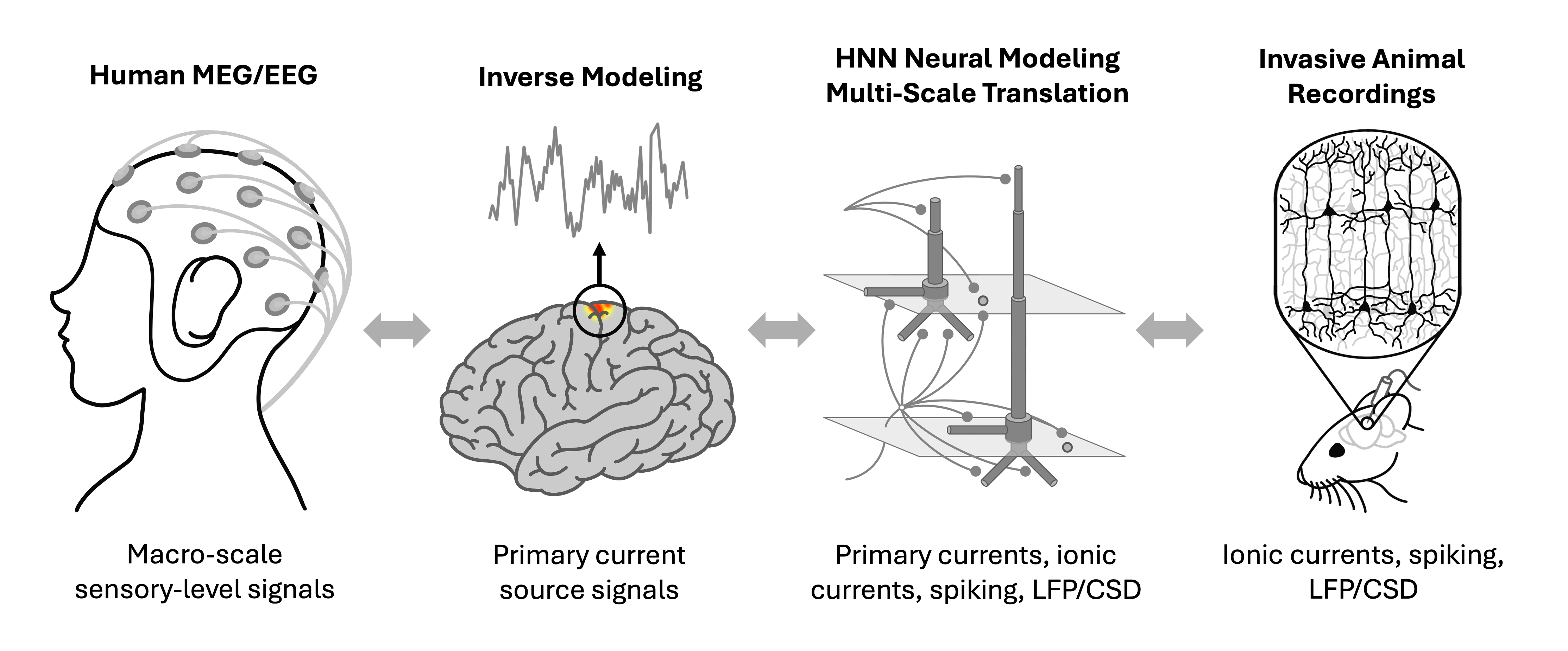
HNN uniquely translates commonly measured human EEG/MEG signals to the wealth of knowledge obtainable in animal studies.
Support our science! If you are willing to volunteer your demographic information using our survey here, this greatly helps to support HNN!
Supporting Institutions







Getting Started
The Human Neocortical Neurosolver (HNN) is an open-source neural modeling tool in Python designed to help researchers/clinicians interpret human brain imaging data. The core of HNN is a large-scale and biophysically-detailed template model of a neocortical column model that simulates primary current dipoles; i.e., the neural sources of the sensor-level EEG/MEG signal.
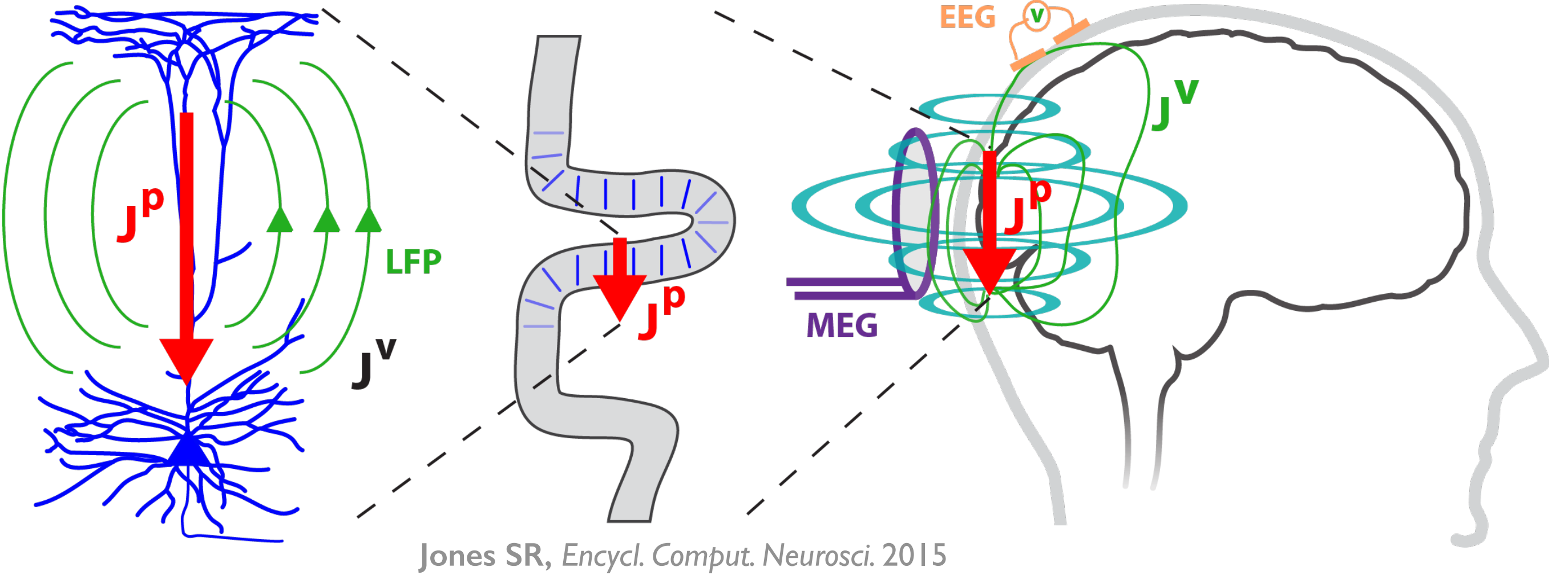
HNN is designed as a starting point for developing and testing predictions on the detailed neural mechanisms contributing to EEG/MEG signals based on the known biophysics of their generation. The multi-scale simulation possible with HNN (layer and cell-specific spiking, LFP/CSD, etc.) facilitates testing of model-derived predictions across multiple recording scales and species.
HNN is designed with workflows to activate the neocortical model with activity from exogenous thalamic and intracortical sources. We provide detailed tutorials on how to study the neural origin of source-localized event-related potentials and low-frequency brain rhythms. Following our workflows, users can quickly develop an intuition of how cells and circuits contribute to macroscale signal generation.
See our
HNN Textbook
to get started with HNN.
The Biophysical Challenge
Electro- and magneto-encephalography (EEG/MEG) are among the most powerful technologies to non-invasively record human brain activity with millisecond resolution. They provide reliable markers of healthy brain function and disease states. A major limitation is that it is often difficult to connect the macroscopic scale-measured signals to the underlying cellular and circuit-level neural generators. This difficulty limits the translation of EEG/MEG studies into novel principles of information processing, or into new treatment modalities for neural pathologies.
HNN is a user-friendly software tool that provides a novel solution to this challenge by giving researchers and clinicians the ability to test and develop hypotheses on the circuit mechanism underlying their EEG/MEG data. The foundation of HNN is a preconstructed biophysically-principled neocortical column model that simulates the electrical activity of the neocortical cells and circuits that generate the primary electrical currents underlying EEG/MEG recordings.
To learn more, see the
Biophysical Modeling
section of our HNN Textbook
ERPs and Rhythms
Our
HNN Textbook
is designed to teach users how to begin studying the origins of ERP and brain rhythms based on our previously-published studies. We provide data, initial parameter and network configurations, and workflows to teach users how to develop and test hypotheses on the neural origin of the most commonly recorded EEG/MEG signals, including event-related potentials (ERPs), Gamma Rhythms (30-80Hz), and low-frequency rhythms including Alpha (7-14Hz) and Beta Rhythms (15-29Hz).
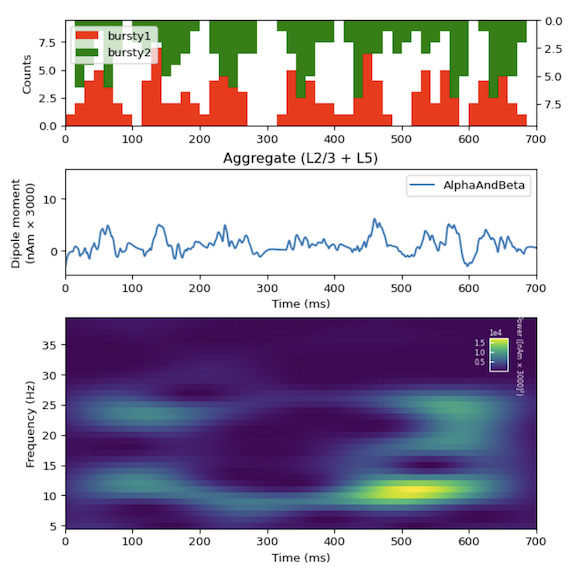
Our simulation examples follow a step-by-step process that serves as a guide on how you can use HNN to develop and test your own hypotheses on the biophysical origins of your recorded signals. You can find our
guide to simulating ERPs here
or our
guide to simulating Alpha/Beta here.
Multi-Scale Visualization
HNN provides easy-to-use interfaces to a wide range of visualization and plotting capabilities across different dimensions and scales of your simulations. At the macro scale, we provide visualizations for simulated and experimental data such as the current dipoles that produce EEG and MEG signals. At the meso scale, we enable simulation and plotting of laminar LFP and CSD output, including the relative position of electrodes with respect to cells. At the micro scale (i.e., on the level of individual cells) we can plot spike rasters, compartmental voltage traces, intracellular state variables like calcium, and many different visualizations of synaptic network connectivity. We provide many tutorials on how to use and visualize in HNN on our code website here.
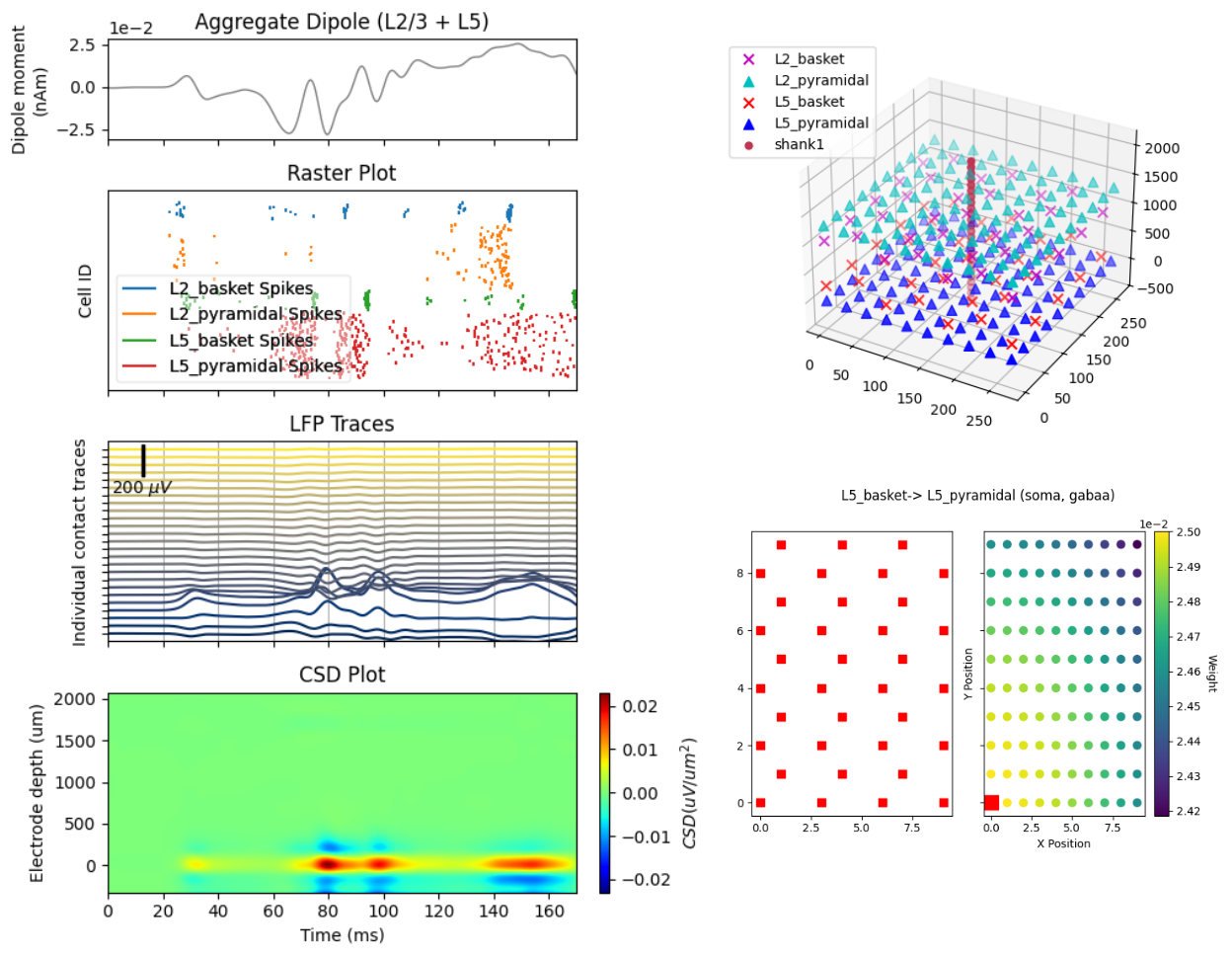
Fitting to Data
HNN provides two ways to fit its pre-tuned neural models to your experimental data:
Hand-tuning: With both the GUI and API, HNN provides all the tools needed for changing parameters to produce new simulations that better match the user's experimental data. HNN is especially powerful for understanding how external drives to the network impact the network and its outputs, including the current dipole that comprises MEG/EEG signals. Manually exploring the simulated parameter space helps users to either test or generate hypotheses tied to specific biological processes, such as statistical changes to inputs, E/I balance, between-layer communication, and much more.
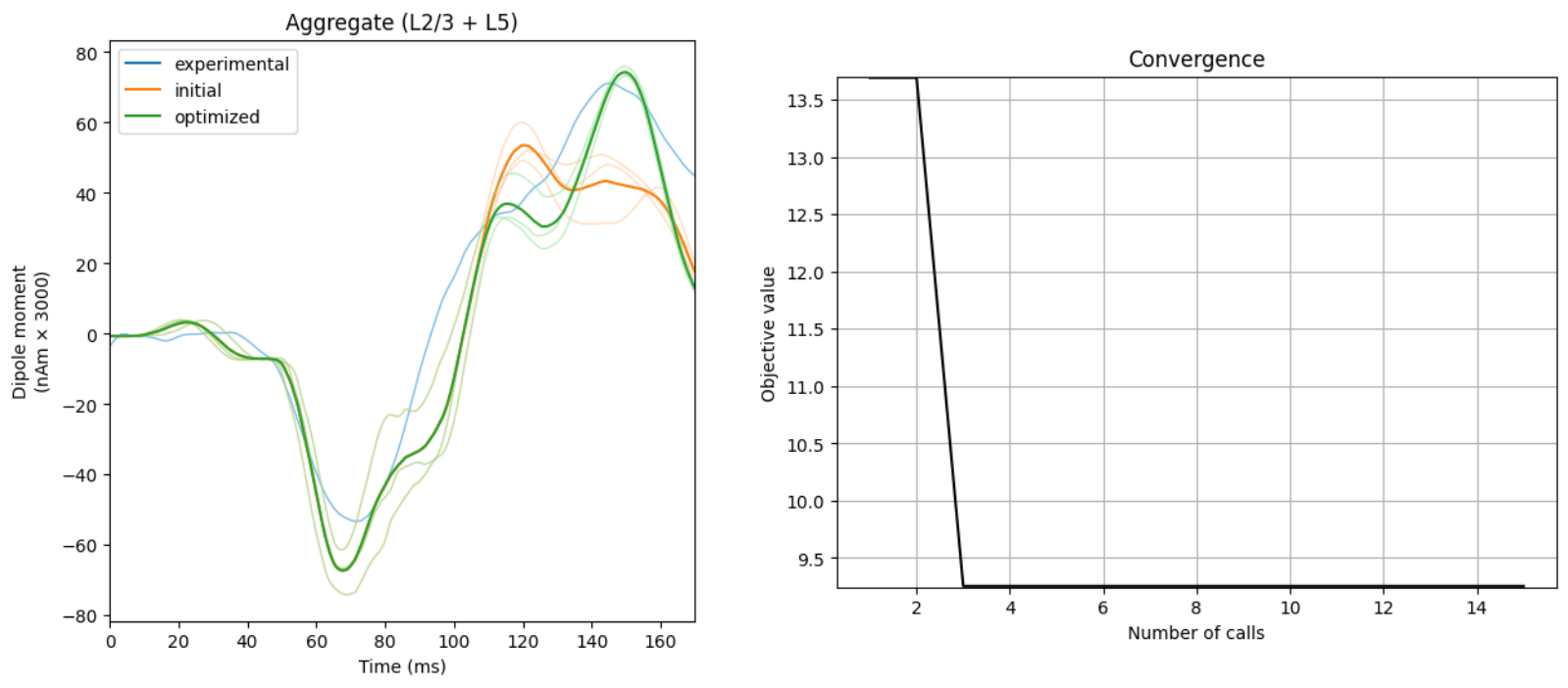
Optimization: Given user-provided constraints, HNN can also minimize the root mean square error (RMSE) between a user-provided experimental dipole signal and simulated dipole signals. Understanding which changes lead to a better match between simulated and experimental data will guide users to novel hypotheses of how the underlying biology works. HNN currently provides support for Bayesian and COBYLA optimization algorithms. See
our Optimization tutorials here
for how to get started using it.
Model Validation
HNN's foundational neocortical column model, referred to as the HNN Template Model (shown below), is a tuned implementation of the computational model in
(Jones et al. 2007).
This model was developed based on canonical principles of neocortical circuitry and contains the minimal circuit elements necessary for generating primary current dipoles and connecting dipole output to layer- and cell-specific neocortical activity.

Building HNN allowed us to test if the cell and network elements thought to be the primary generators of ERPs, alpha rhythms, beta rhythms, gamma rhythms, and other activity could produce macro-scale current dipole signals that were in agreement with our recorded data. While the literature guided us to a starting point that constrained the investigation, the exact parameters to use were unknown. By directly comparing the current dipole output of the model to source-localized experimental data, we were able to titrate parameters (both through hand tuning and algorithmic parameter estimation) to produce a close fit to the source waveform (i.e., using small root mean squared error (RMSE)). Once a close fit to the current dipole waveform was established, the model provided a number of testable predictions on the underlying cell and circuit activity; e.g. exogenous input timing and strength, cell spiking activity, LFP/CSD, etc.
For more detail on how our Template Model was validated, see the
Model Assumptions
section of our HNN Textbook. For publications that have used our model, see
our Publications page.